ASFv: Genetic variation and evolution

African Swine Fever can lead to high levels of pig mortality. What, however, is known about the virus itself and its genetic origin? In total, 24 different genotypes have been described. Only genotypes I and II have spread to countries outside the African continent.
African Swine Fever (ASF) is a highly contagious haemorrhagic disease that causes high mortality rates in swine production systems worldwide. The disease is caused by ASF virus (ASFv), and it was first reported in the 19th century and then continued to spread in Africa and other parts of the world.
Risk of transmission and spread of ASFv
The growing demand for pork and the concomitant increase in transboundary movement of pigs and pork products increase the risk of transmission and spread of ASF and pose a major challenge to the pig industry. Factors such as the extreme genetic and antigenic diversity of the virus and poor cross-protective immunity are the main obstacles to developing a safe and efficacious vaccine against ASF.
Understanding genotype circulation
Different genotypes of ASFv with varying virulence have been associated with different outbreaks worldwide, and understanding genotype circulation is essential for ASF prevention and control strategies. Researchers classify the virus into 24 genotypes (categorised I–XXIV) using several specific genetic targets including p72 (B646L), CVR (B602L) and p54 (E183L). This article will discuss ASFv biology, the importance of genotyping, geographic distribution of the virus’ genotypes, and ASFv serogroups.
African Swine Fever virus biology
ASFv has a large linear double-stranded DNA with multiple envelopes and is the only virus classified under the Asfarviridae family and Asfivirus genus. The genome is about 170–190 kb and is divided into the left variable region (3,848 kb), the central conserved region (about 125 kb) and the right variable region (13–22 kb). The ASFv encodes 150–165 proteins with essential functions in replication and evasion of host immune responses. Various ASFv strains are considerably different in important positions including the multigene family in the left variable region, the central variable region in the central conserved region and the EP402R expressing CD2v protein genes.
The importance of genotyping
Genotyping ASFv during outbreaks is essential to establish the origin of the virus, to distinguish between closely related strains and to understand ASFv diversity and evolution. Genetic variation analysis of ASFv is performed by sequencing specific genetic markers.
One of the markers used for relatively quick and easy typing of ASFv is the carboxy terminal end of the p72 gene (B646L). That method identifies the origin of a new outbreak and categorises ASFv into the 24 genotypes; however, it does not distinguish closely related viruses.
The central variable region of the B602L gene is another marker used for intragenotypic differentiation, which identifies various subgroups of ASFv. The p54 (E183L) is another commonly used genetic target to assess ASFv genotype, which increases the intragenotypic resolution of standard p72 genotyping. There are also various other established genotyping methods to distinguish between evolutionarily similar isolates of ASFv.
The development of next-generation sequencing and third-generation sequencing techniques enabled the reduction of genome sequencing cost, and hence made large-scale sequencing for large double-stranded DNA virus genomes feasible. Later, phylogenetic analysis based on the 440 bp-length partial sequence of the approximately 1,500 bp-length B646L gene revealed 24 genotypes of ASFv. All these genotypes were found to be endemic in Africa.
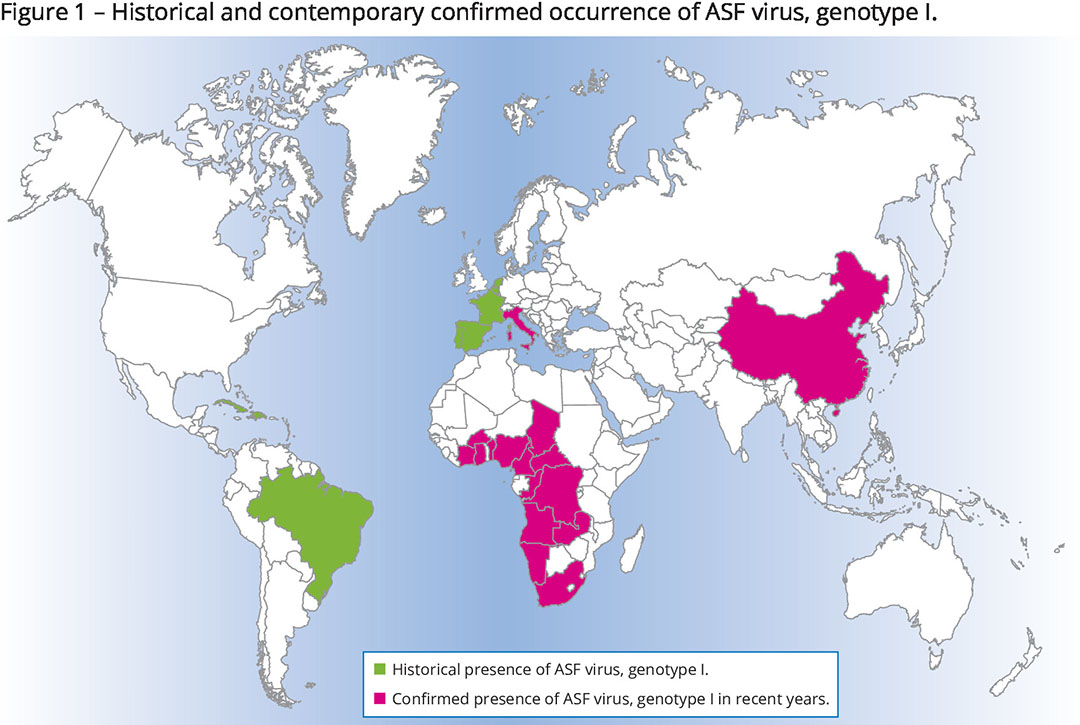
Genotype I
2 of the genotypes, genotype I and genotype II, have already caught the world’s attention as they have managed to expand outside the African continent. As shown in Figure 1, genotype I was originally a problem in western Africa, but from the 1950s it crossed the Mediterranean to Spain and Portugal where it remained present until the mid-1990s. Incidental excursions north to France, Belgium and the Netherlands have been reported, as well as to various countries in Central and South America. Currently, all that is left of that wave is the fact that the virus is still endemic among pigs in Sardinia, an island belonging to Italy, a situation that has been monitored well. Interestingly, genotype I was recently also confirmed in China.
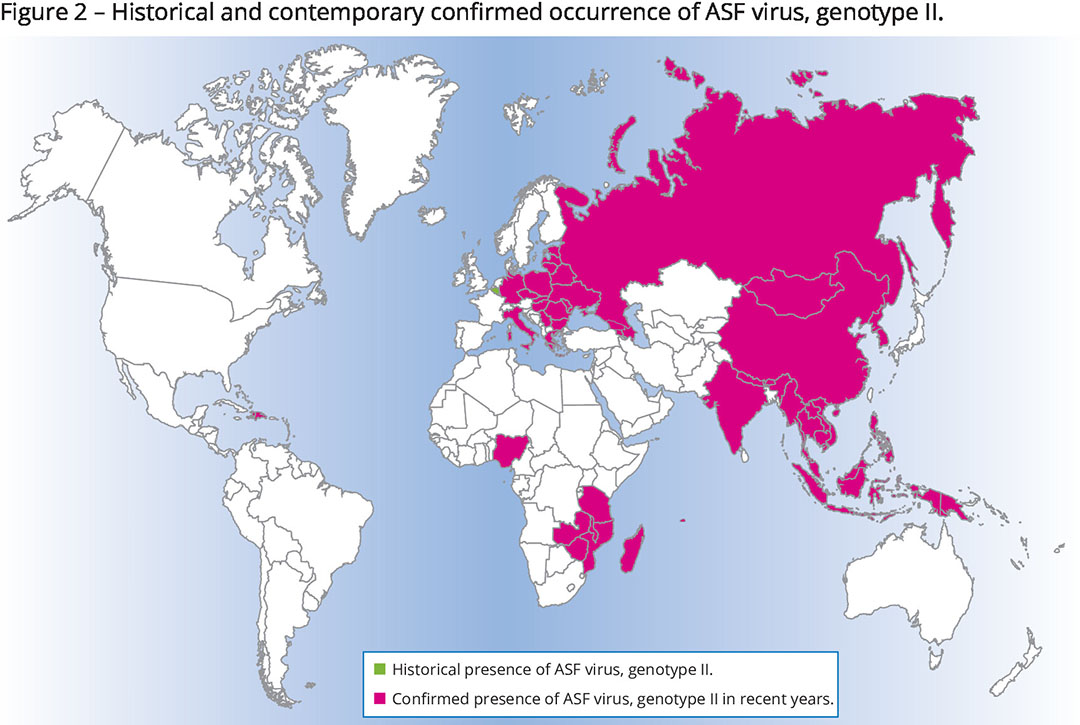
Genotype II
Until 2007, ASFv genotype II was mostly a problem for eastern Africa. In that year, it was also confirmed in the Caucasus region of Georgia. Figure 2 shows how it then progressively spread to Eastern and Central Europe to reach a total of (currently) 20 countries in this region, including countries as far west as Germany and Italy. Only Belgium and the Czech Republic managed to eradicate an outbreak of ASFv, genotype II, but late in 2022 the virus resurfaced in wild boar in the Czech Republic.
The wave also spread east: the first occurrence of ASFv in Asia was reported in 2018 in China and Vietnam, leading to millions of pig deaths. It then spread to 19 Asian countries as far east as Papua New Guinea. In 2021, ASFv genotype II appeared in the Americas with outbreaks detected in the Dominican Republic and Haiti.
Behaviour in the field
According to a research study by Encheng Sun and colleagues (2021) from Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, the genotype I of ASFv that emerged in China had lower virulence than ASFv genotype II and caused milder onset of infection and chronic disease. However, genotype II of ASFv isolated in China causes very acute disease with near 100% mortality.
In 2020, lower virulent genotype II of ASF viruses emerged in China due to natural mutations in the genomes of the highly virulent viruses. These natural mutants showed lower virulence and high transmissibility, caused chronic and persistent infections in pigs, but were continuously shed via the oral and rectal routes at a low level, which causes more difficulties and challenges for the early diagnosis and control of ASF in China.
Based on reviewing multiple studies, both genotypes I and II of ASFv can have low, moderate and high virulence isolates, and the symptom in pigs depends on the virulence level of each isolate.
Not very much is known about the other genotypes. Most literature explains where genotypes III–XXIV are isolated (in Africa) and the genotyping process, but other than that, not much information is available.
Serogroups of ASFv
Learning about more common serotypes improves understanding of ASF and the natural history of all the ASFv strains. Furthermore, the correlation between currently established ASFv genotypes and viral cross-protection is not precisely clear. Thus, having knowledge about the relationship of the established genotypes with serologic classification is essential for vaccine development. In vaccine design and development, consideration should be given to the fact that viruses within a serotype provide cross-protection from challenge with viruses of the same serotype.
ASFv is one of the few viruses without neutralising antibodies. Using a haemagglutination inhibition test to evaluate serological cross-reactivity with different ASFv isolates, eight ASFv antigenic serogroups have been identified. More serogroups may be detected if additional parameters such as haemadsorption density, which is the number of red blood cells per infected cell, and red blood cell contact maps are used to refine the classification of ASFv isolates.
Focus for future research
Understanding new genetic markers involved in ASFv virulence is essential to identify high-risk routes of cross-border spread and to develop control strategies against the virus. Further research is required to discover new ASFv genetic markers connected to the evolution of ASFv isolates in parts of the world where the disease is endemic. In addition, further research focusing on the molecular factors affecting asymptomatic ASFv infection in African suids is needed to improve understanding of virus transmission routes, virus presence and continued existence in diseased tissues, and a better characterisation of carriers on potential clinical activation.
Identifying the virulence factors and mechanisms of pathogenesis of ASF improves diagnosis and early detection of severe or rapidly spreading outbreaks of the virus on farms and fields. Therefore, establishing and characterising genomic markers associated with ASFv virulence can help in developing more suitable diagnostic approaches in infected animals.
Concluding remarks
Specific genetic targets including p72 (B646L), CVR (B602L) and p54 (E183L) are used to identify various genotypes of ASFv and to track the virus in a particular region. As a result, 24 ASFv genotypes have been identified based on the p72-encoding (B646L) gene. The p72 genotyping traces the source of the virus at the molecular level and assists with understanding the potential transmission routes and possible modes of transmission.
The CVR (B602L) and p54 (E183L) are used to predict molecular epidemiological changes and evolution of ASFv. Furthermore, application of in-depth ASFv genomes and typing technology –especially whole-genome sequencing and cross-protection experiment data – clarifies biology, evolution and genetic characteristics of ASFv.
References available upon request.
 Beheer
Beheer

![Photo: [1:20 PM] Vincent ter Beek](jpg/img_3000x2000asf-pigcopy-389x260.jpg)





 WP Admin
WP Admin  Bewerk bericht
Bewerk bericht